

Fungal cell walls are rigid and made up of complex polysaccharides called chitin and glucans. Chitinase is an hydrolytic enzyme that degrades chitin and is secreted as a natural defense against phytopathogenic fungi. Chitinases are naturally produced by various organisms, viz. fungi, bacteria, yeats, plants, actinomycetes, arthropods, and humans. Chitinases are classified into three families: GH18, GH19 and GH20 glycosidic hydrolase families. Many GH18 family chitinases contain a catalytic domain (CD) which has a Triosephosphate Isomerase barrel (TIM barrel) structure and an additional Chitinase Insertion Domain (CID). Often, separate chitin-binding domains (CBDs) are present in the carboxy-terminal of the proteins. Studies have shown enhanced activities for chimeric chitinases with additional domains than the native structure. Here, we proposed to develop hybrid bacterial and plant chitinases by fusing domains from different source organisms.
We designed two chimeric bacteria chitinases from three bacterial sources: Pseudoalteromonas sp DL-6, Amycolatopsis orientalis, and Serratia marcescens.
First chimeric combination: We replaced the catalytic domain of Serratia marcescens with the catalytic domain of Pseudoalteromonas following the basic structure of Serratia chitinase.
Amino Acids in the beginning of Serratia ChiB natural sequence — CBD of S. marcescens Chi B — CD of Pseudoalteromonas Chi A — Chi B natural linker between CD and CBD of S. marcescens — CBD of S. marcescens
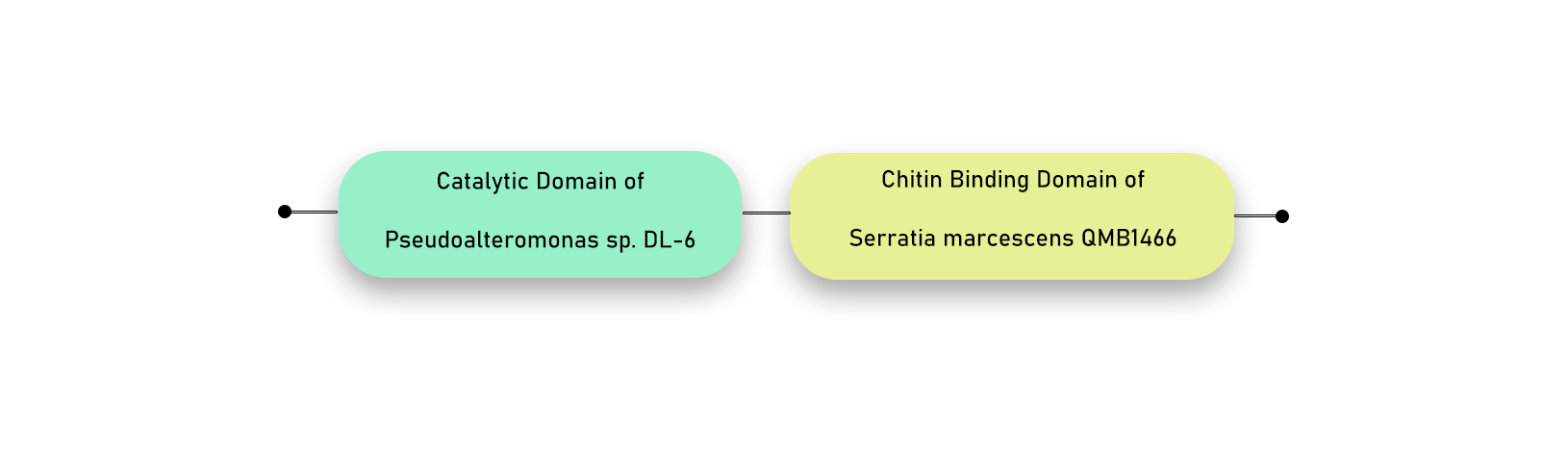
Figure 1. Unannotated image
Second chimeric combination: We chose chitinase from Amycolaptosis orientalis which lacks a chitin-binding domain. So we added the chitin-binding domain of S. marcescens Chi B in the C-terminal of the Amycolatopsis orientalis chitinase.
Chitinase seq. of Amycolatopsis orientalis — CBD of S. marcescens Chi B
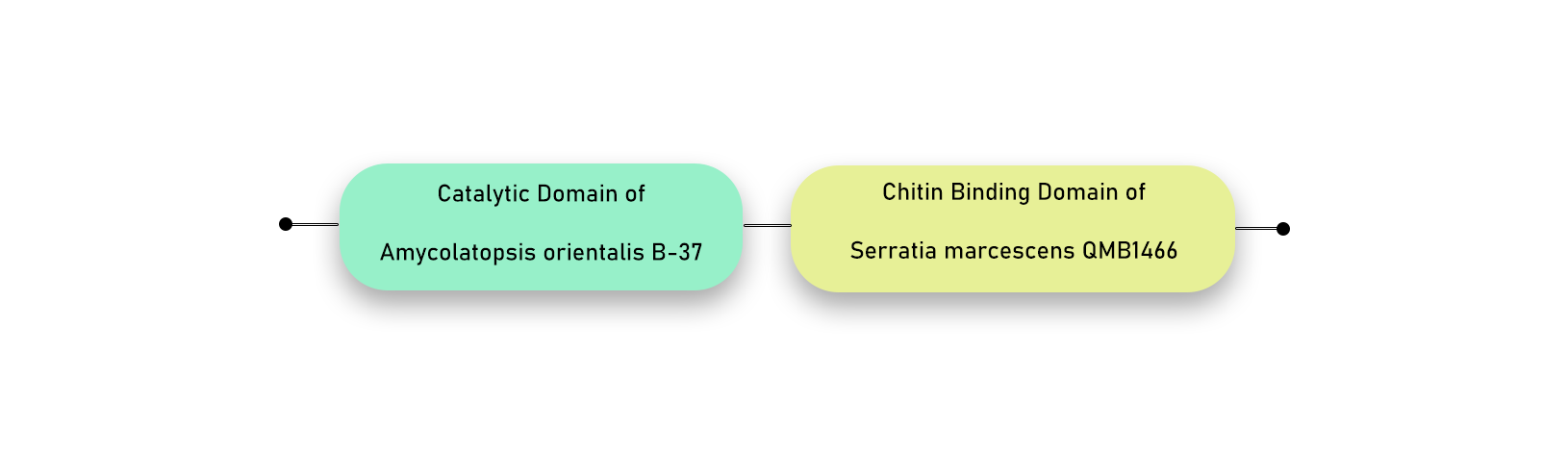
Figure 1. Unannotated image
Apart from bacterial chitinases, we chose plant chitinases based on suggestions from structural biology experts since these chitinases would have evolved with more efficient chitinolytic activity. We designed two chimeric plant chitinase combinations using chitinases from two sources: Triticum aestivum (Wheat) and Hordeum vulgare (Barley). The cloning and the succeeding downstream assay experiments could not be performed due to lack of time. Both of these plant chitinases show efficient chitinolytic activity and retain activity at body temperature and pH range.
First chimeric combination: We joined the sequences of both barley and wheat chitinases using common (GGGS)\(_3\) linkers. Wheat chitinase doesn’t have a chitin-binding domain, so we added barley CBD in its C-terminal in the combination.
AA seq before Barley CBD — Barley CBD — Barley CD — AA after CD barley — (GGGS)\(_{3}\) — AA seq before wheat CD — Wheat CD — natural linker from barley — Barley CBD
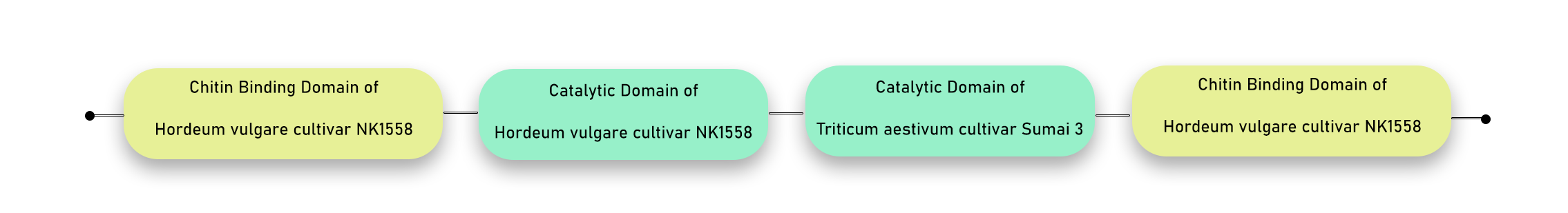
Figure 1. Unannotated image
Second chimeric combination: We created a chimeric chitinase with a wheat catalytic domain flanked by two barley chitin-binding domains.
AA before barley chitinase — Barley CBD — natural linker from barley — Wheat CD — natural linker from Barley — Barley CBD
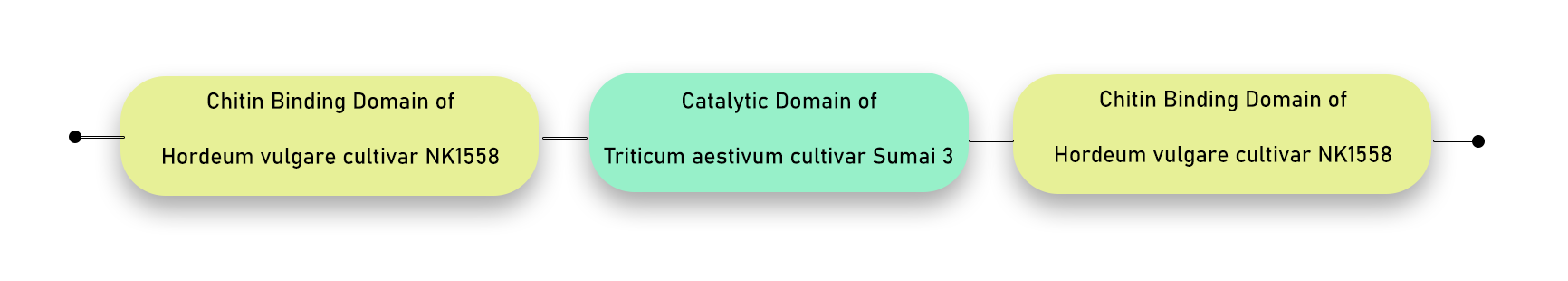
Figure 1. Unannotated image
The sequences that we created are codon-optimized in order to clone and express them in E. coli.